Enhanced QIAseq DIRECT SARS-CoV-2 Kit for Illumina MiSeq
Padmini Ramachandran, Tamara Walsky, Amanda Windsor, Maria Hoffmann, Christopher Grim
Abstract
This SARS-CoV-2 targeted, a rapid viral RNA library preparation kit compatible with Illumina sequencers, provides an approximately five hour turn-around time from viral RNA sample to sequencing. The kit includes cDNA synthesis, SARS-CoV-2 enrichment, and library amplification and indexing reagents. The Enhanced protocol removes the need for normalization and quantification prior to amplification and indexing. RNA input is 5 μl, regardless of SC2 viral load in the sample extract. Amplification is performed using two primer pools that produce overlapping amplicons that are approximately 250 bp in size; as a result, no fragmentation is necessary prior to sequencing on Illumina platforms. 10 bp unique dual indexes (UDIs) are provided in the kit, and with a cycle read length of 149 for each paired-end read, the sequencing time is just over 28 hours.
Steps
cDNA Synthesis Procedure
Thaw template RNA on ice. Gently yet thoroughly mix, then briefly centrifuge to collect residual liquid from the sides of the tubes and return to ice.
While keeping reagents on ice, prepare the cDNA reaction according to the table below. Briefly centrifuge, “gently yet thoroughly” vortex to mix, and centrifuge briefly again. For multiple reactions, prepare the calculated volume for all reactions, plus 10%.
| A | B |
|---|---|
| EZ Reverse Transcriptase | 1 |
| Multimodal RT Buffer, 5x | 4 |
| Nuclease-free water | 8 |
| RNase Inhibitor | 1 |
| RP Primer | 1 |
| Template RNA | 5 |
| Total volume | 20 |
cDNA synthesis incubation. Incubate as described below.
| A | B | C |
|---|---|---|
| Step | Temperature (C) | Incubation time (min) |
| 1 | 25 | 10 |
| 2 | 42 | 50 |
| 3 | 85 | 5 |
| 4 | 4 | Hold |
Proceed to“ SARS-CoV-2 Enrichment Procedure”. Alternatively, the samples can be stored at−30 to–15°C in a constant-temperature freeze.
SARS-CoV-2 Enrichment Procedure
While keeping reagents on ice, set up Pool 1 and Pool 2 reactions for each sample, as described below. Briefly centrifuge, “gently yet thoroughly” vortex to mix, and centrifuge briefly again.
| A | B | C |
|---|---|---|
| Component | Pool 1: Volume/reaction (µl) | Pool 2: Volume/reaction (µl) |
| cDNA from “Protocol: Enhanced QIAseq DIRECT” | 8 | 8 |
| DIRECT SARS-CoV-2 Pool 1 | 2 | --- |
| DIRECT SARS-CoV-2 Pool 2 | --- | 2 |
| UPCR Buffer, 5x | 5 | 5 |
| QN Taq Polymerase | 1 | 1 |
| Nuclease-free water | 9 | 9 |
| Total volume | 25 | 25 |
Important: For samples with a broad/unknown range of Ct values (i.e., above and below Ct = 32) incubate as described in Table 9.1. For samples with Ct > 32, also incubate as described in Table 9.1. For samples with Ct value < 32, incubate as described in Table 9.2.
Target enrichment cycling: Samples with broad/unknown range Ct values and samples with Ct > 32
| A | B | C | D |
|---|---|---|---|
| Step | Time | Temperature (°C) | Number of cycles |
| Heat activation | 2 min | 98 | 1 |
| Denaturation | 20 s | 98 | 4 |
| Annealing/extension | 5 min | 63 | |
| Denaturation | 20 s | 98 | 29 |
| Annealing/extension | 3 min | 63 | |
| Final Hold | ∞ | 4 | Hold |
Target enrichment cycling: Samples with Ct < 32
| A | B | C | D |
|---|---|---|---|
| Step | Time | Temperature (°C) | Number of cycles |
| Heat activation | 2 min | 98 | 1 |
| Denaturation | 20 s | 98 | 4 |
| Annealing/extension | 5 min | 63 | |
| Denaturation | 20 s | 98 | 20 |
| Annealing/extension | 3 min | 63 | |
| Final Hold | ∞ | 4 | Hold |
After amplification, combine the entire contents of “Pool 1” and “Pool 2” PCR reactions for each biological sample into a single well of a plate, giving a volume of 50 µl.
Add 50 µl QIAseq Beads to each 50 µl combined sample. Briefly centrifuge ,“gently yet thoroughly” vortex to mix, and centrifuge briefly again.
Incubate for 5min at room temperature.
Centrifuge the plate until the beads are pelleted (2 min), and then place the plate onto a magnetic rack. After the solution has cleared (2 min or longer), carefully remove and discard the supernatant.

With the plate still on the magnetic stand, add 200 µl of 80% ethanol. Wait 1 min and carefully remove and discard the wash. Repeat the wash, for a total of 2 ethanol washes. Remove excess ethanol as much as possible.
With the plate still on the magnetic stand, air-dry at room temperature for 4–8 min (until the beads start to crack and pellet loses its shine).

Remove the plate from the magnetic stand and elute the DNA from the beads by adding 30 μl nuclease-free water. “Gently yet thoroughly” vortex (triturate if necessary) to mix, briefly centrifuge, incubate for 2 min.
Centrifuge the plate until the beads are pelleted (2 min), and then return the plate to the magnetic rack until the solution has cleared.
Transfer 28 μl to a clean plate. This is now “enriched SARS-CoV-2”.
Important Notes:
-
We are adding tails ONLY to SARS-CoV-2 sequence in the subsequent Library Amplification and Indexing Procedure, and that will keep all the contaminants at bay when you perform your final amplification.
-
So, after you transferred the 28 µl of supernatant from your elution, this is a good time to get a feel for what you have if you want to do a mid-protocol check.
-
Use a high sensitivity Qubit (for example) and check the concentration of the enrichment reactions after bead cleanup.
-
With wastewater samples anything that is 1ng/µl and above (even down to 0.5ng/µl with high Ct values) is a success and should definitely move on to final amplification!
Proceed to “Library Amplification and Indexing Procedure”. Alternatively, the samples can be stored at –30 to –15°C in a constant-temperature freezer.
Library Amplification and Indexing Procedure
For samples with a broad/unknown range of Ct values (i.e., above and below Ct = 32) incubate as described in Table 23.1. For samples with Ct > 32, also incubate as described in Table 23.1. For samples with Ct value < 32, incubate as described in Table 23.2.
Library amplification/indexing cycling conditions: Samples with broad/unknown range Ct values and samples with Ct > 32
| A | B | C | D |
|---|---|---|---|
| Step | Temperature (°C) | Time | Number of cycles |
| Initial denaturation | 98 | 2 min | 1 |
| 3-step cycling | |||
| Denaturation | 98 | 20 s | |
| Annealing | 60 | 30 s | 11 |
| Extension | 72 | 30 s | |
| Final extension | 72 | 1 min | 1 |
| Final Hold | 4 | ∞ | Hold |
Library amplification/indexing cycling conditions: Samples with Ct < 32
| A | B | C | D |
|---|---|---|---|
| Step | Temperature (°C) | Time | Number of cycles |
| Initial denaturation | 98 | 2 min | 1 |
| 3-step cycling | |||
| Denaturation | 98 | 20 s | |
| Annealing | 60 | 30 s | 14 |
| Extension | 72 | 30 s | |
| Final extension | 72 | 1 min | 1 |
| Hold | 4 | ∞ | Hold |
On ice, prepare the library amplification and indexing reaction according to Table in Step 25. Briefly centrifuge, “gently yet thoroughly” vortex to mix, and centrifuge briefly again.
Reaction mix for library amplification and indexing
| A | B |
|---|---|
| Component | Volume/reaction |
| “Enriched SARS-CoV-2” sample | 24 μl |
| Index from QIAseq DIRECT UDI index plate (A, B, C, or D) Plate | 2 μl |
| Nuclease-free water | 12 μl |
| UPCR Buffer 5x | 10 μl |
| QN Taq Polymerase | 2 μl |
| Total reaction volume | 50 μl |
Transfer the plate to the thermal cycler and start the program.
Once PCR is complete, add 40 μl of resuspended QIAseq Beads to each 50 μl reaction. Briefly centrifuge, “gently yet thoroughly” vortex to mix, and centrifuge briefly again.
Incubate the mixture for 5 min at room temperature.
Centrifuge the plate until the beads are pelleted (2 min), and then place the plate onto a magnetic rack. After the solution has cleared (2 min or longer), carefully remove and discard the supernatant.
With the plate still on the magnetic stand, add 200 μl of 80% ethanol. Wait 1 min and carefully remove and discard the wash. Repeat the wash, for a total of 2 ethanol washes. Remove excess ethanol as much as possible.
With the plate still on the magnetic stand, air-dry at room temperature for 4–8 min (until the beads start to crack and pellet loses its shine).

Remove the plate from the magnetic stand and elute the DNA from the beads by adding 25 μl nuclease-free water. Vigorously vortex (triturate if necessary) to mix, briefly centrifuge, and incubate for 2 min.
Centrifuge the plate until the beads are pelleted (2 min), and then return the plate to the magnetic rack until the solution has cleared (2 min or longer).
Transfer 23 μl to a clean plate. This is the “SARS-CoV-2 library”. If not proceeding immediately, the sample can be stored at –30 to –15°C.
Quantify the library using Qubit HS assay kit.
Quantification, Normalization and Pooling of Libraries
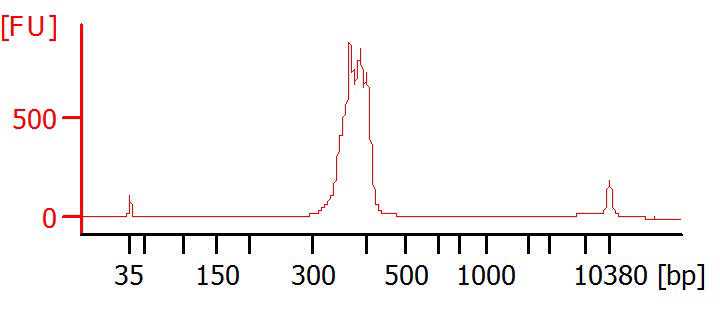
Using the attached Post library worksheet, calculate the nM based on the Qubit concentration (ng/ul) and the Tape station (bp) profile. Normalize the “SARS-CoV-2 library”, to 4 nM. Quantify the final pool at the end.
Proceed to loading on to MiSeq (150x 150 cycle) using the attached Sample Sheet. Alternatively, the purified “SARS-CoV-2 library” can be safely stored at –30 to –15°C in a constant-temperature freezer until ready to use for sequencing.SampleSheet.csv