Engineering brain assembloids to interrogate human neural circuits
Yuki Miura, Min-Yin Li, Omer Revah, Se-Jin Yoon, Genta Narazaki, Sergiu Pasca
human neural circuits
brain assembloids
viral labeling
retrograde tracing
3D live imaging of axon projection
optogenetics
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
The development of neural circuits involves wiring of neurons locally following their generation and migration, as well as establishing long-distance connections between brain regions. Studying these developmental processes in the human nervous system remains difficult because of limited access to the tissue that can be maintained as functional over time in vitro. We have previously developed a method to convert human pluripotent stem cells into regionalized neural organoids that can be fused and integrated to form assembloids and study neuronal migration. In this protocol, we describe approaches to model long-range neuronal connectivity in human brain assembloids. We present how to generate 3D spheroids resembling specific domains of the nervous system and then how to integrate them physically to allow axonal projections and synaptic assembly. In addition, we describe a series of assays including viral labeling and retrograde tracing, 3D live imaging of axon projection and optogenetics combined with calcium imaging and electrophysiological recordings to probe and manipulate the circuits in assembloids. We anticipate that these approaches will be useful in deciphering human-specific aspects of neural circuit assembly and in modeling neurodevelopmental disorders with patient-derived cells.
Before start
CRITICAL: Before starting hPS cells and the long-term organoid/assembloid cultures, carefully cleaning the incubator is important to avoid potential contamination. Because most culture plates will be in the incubator for several hundred days, the surface of the incubator and culture dishes should be regularly cleaned. The amount of water in the incubator should be monitored frequently to maintain the required humidity. All cell culture-related procedures should be performed in a sterile environment with biological safety cabinets and sterilized materials. The biological safety cabinets should be regularly certified.
Steps
(B) Generation of hStrSs
From day 6, replace the medium daily with NM supplemented with 50ng/ml activin A and 2.5micromolar (µM) IWP-2. Change the medium daily from day 6 to day 11.
Viral labeling of neural spheroids ● Timing 3 d
For labeling neural spheroids, dilute 0.5µL of virus solution (recommended virus particle volumes are listed in Table 1) in 200µL of NM and add mixed NM to a 1.5mL Eppendorf tube. Then, transfer one to three spheroids in the tube containing NM and viruses. We note that transduction efficiency depends on the target cell types, the promoter, stage of differentiation and brain region. The tube can be placed in an incubator at 37°C, 5% CO2.
Generating cortico-striatal assembloids ● Timing 3–4 d
(C) Cryosection and immunostaining ● Timing 6–7 d
(i) When ready to perform immunocytochemistry, transfer spheroids to a 1.5mLmicrocentrifuge tube by using a cut P1000 tip. Gently remove the medium and add 500µL to 1mLof cold 4% PFA/PBS (or warm 4% PFA/4% sucrose/PBS). Leave spheroids at4°C 24h 0m 0s.
(ii) After fixation, gently remove PFA and wash with PBS at 4Room temperature.
(iii) Add 1mL of 30% (wt/vol) sucrose/PBS for cryopreservation. Leave spheroids or assembloids at 4°C until they sink to the bottom of tube (≥24h 0m 0s to 48h 0m 0s).
(iv) Prepare square disposable molds that fit with the sample size and number and fill the molds with embedding solution (1:1, OCT/30% sucrose; see Reagent setup).
(v) Transfer spheroids from tubes into the mold by using a P1000 cut pipette tip. Place 4–10 spheroids in the mold. If spheroids move while embedding, use a P20 tip to gently place them in the embedding solution.
(vi) Place the spheroids in a cryomold 4On ice for 0h 20m 0s and allow the spheroids to sink to the bottom of the mold.
(vii) Snap-freeze the spheroids by placing the mold directly on dry ice. Once completely frozen, store molds at-80°C.
(viii) When ready for sectioning, transfer the mold from -80°C to the cryostat chamber at -20°C for ~0h 30m 0s before sectioning.
(ix) Remove the block from the mold, paste the specimen on the stage by using OCT and section by using standard techniques. Use a brush to prevent crumbling of the sections. Spheroids and assembloids can be cryo-sectioned at 10 µm to 30 µm thickness, depending on the purpose of the experiments.
(x) Collect sections on Superfrost Plus slides. Slides can be stored at -20°Cto -80°Cuntil ready to immunostain.
(xi) For immunostaining, remove the slide from the freezer and leave at 80Room temperature to thaw for 0h 5m 0s to 0h 10m 0s.
(xii) Wash the slide three times with PBS to remove excess OCT/sucrose on the slides.
(xiii) Remove PBS and block for 1h 0m 0sat80Room temperature with 10% normal donkey serum (NDS), 0.3% Triton- X and 0.1% BSA in PBS. Parafilm can be placed on top of the slide glass to avoid drying of the antibody solution. Alternatively, a hydrophobic PAP pen can be used to draw circles around the sections, and staining can be performed inside these circles.
(xiv) Perform the immunostaining steps in a humidified chamber to avoid drying of sections.
(xv) Add the primary antibodies diluted in antibody solution containing 2% NDS and 0.1% Triton-X in PBS and keep at 4°C``1h 0m 0s.
(xvi) The next day, perform three 0h 5m 0s PBS washes at 4Room temperature.
(xvii) Add secondary antibodies diluted in antibody solution for 1h 0m 0s at 4Room temperature Parafilm can be placed on top of the slide glass to avoid drying.
(xviii) Perform three 0h 5m 0s washes at4Room temperature with PBS.
(xix) Add Hoechst in PBS (1:10,000 dilution) for 0h 5m 0sto 0h 7m 0sat 4Room temperature.
(xx) Aspirate the Hoechst solution and wash twice with PBS.
(xxi) Aspirate PBS and mount the sections by adding one drop of Aquamount solution on top of sections and by placing a coverglass on top.
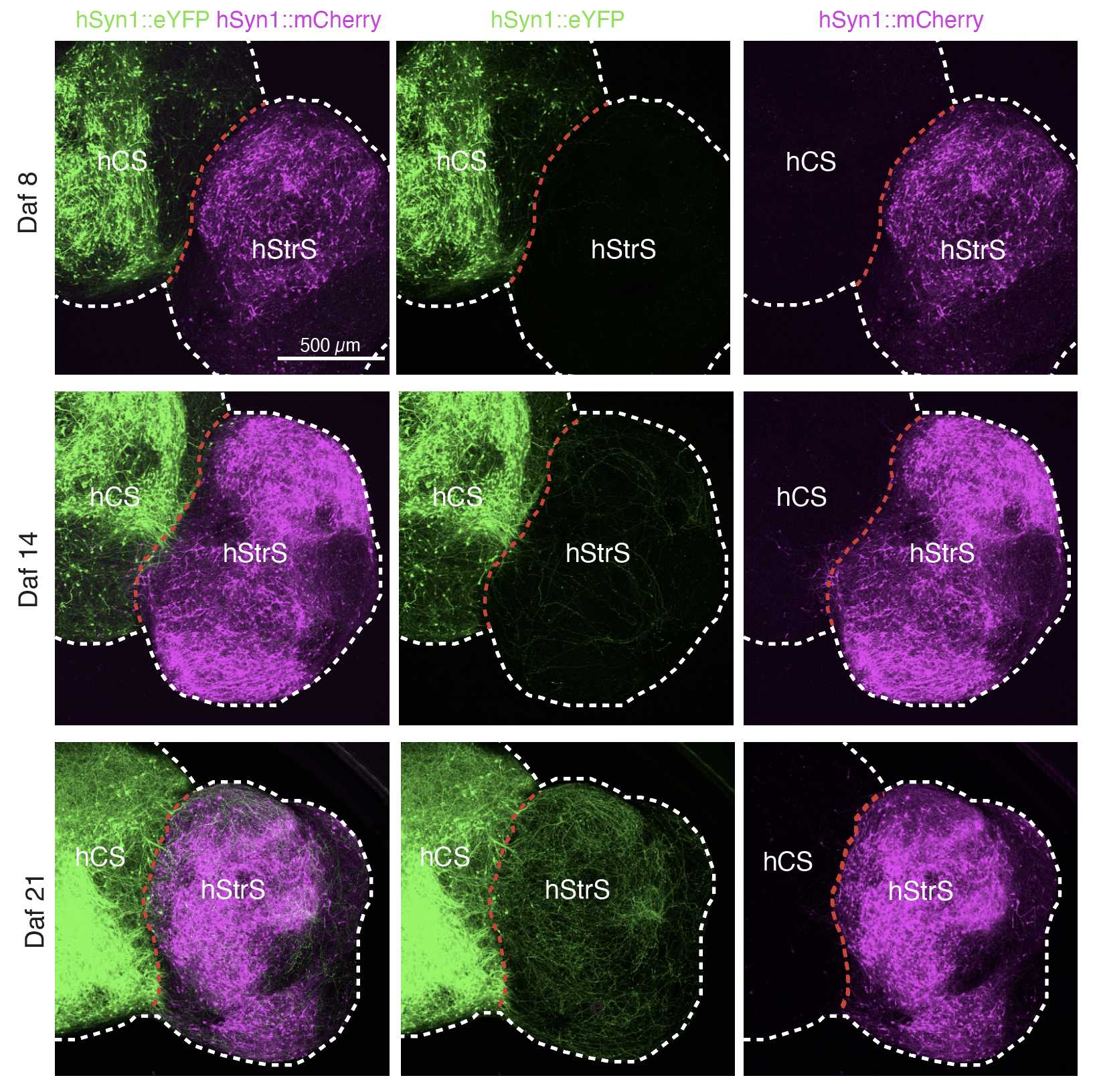
(xxii) Use a fluorescent microscope to image (Fig. 2j, k and Fig. 3f–i).
.
Maintenance of hPS cells in feeder-free conditions Timing 4–6 d
Culture hPS cells on human recombinant vitronectin with Essential 8 medium. When hPS cell cultures reach ~80% confluency, passage hPS cultures by using0.5millimolar (mM) EDTA in PBS.
Regularly check hPS cell cultures for mycoplasma (e.g., PCR) and chromosomal abnormalities (e.g., SNP or comparative genomic hybridization arrays).
Preparing vitronectin-coated dishes
Prepare the vitronectin-coated plates at 37Room temperature and pre-warm the required volume of Essential 8 medium at 37Room temperature.
To coat a well of a six-well plate, dilute 60µLof vitronectin in 6mL (1:100) of DPBS (without calcium and magnesium), add 1mLof diluted vitronectin solution to the well and incubate at 37Room temperature for 1h 0m 0s.
Passaging hPS cells
Aspirate culture medium from the hPS cells plate and rinse with2mL of DPBS (without calcium and magnesium).
Add 1mLof 0.5millimolar (mM)EDTA in DPBS to the well of a six-well plate and incubate at 37Room temperature for 0h 7m 0s.
Aspirate the EDTA solution and add 1mL of pre-warmed complete Essential 8 medium per well of a six-well plate.
Remove cells from the well by gently squirting medium and pipetting the colonies with either a 5mL serological pipette or P1000 tips with a pipette.
Aspirate the residual vitronectin solution from the pre-coated plate.
Add 2mL of pre-warmed Essential 8 medium to each well of a coated six-well plate. Mix the cell suspensions by pipetting and transfer the appropriate volume of cell suspension into each well containing pre-warmed complete Essential 8 medium according to the desired split ratio (1:6 to 1:20 ratio, depending on the confluency and growth rate of individual hPS cell lines).
Move the plate in several quick, figure-of-eight-like movements to disperse cells.
Gently place the plate in a 37°C, 5% CO2 incubator.
Perform a medium change after every passage (every18h 0m 0sto24h 0m 0s), except for the day after passage, when confluency of cells is low.
Preparation of hPS cells for 3D neural differentiation
Passage hPS cells from a six-well plate to 10-cm plates and grow to ~80–85% confluency before the day of aggregation.
48h 0m 0s and 24h 0m 0sbefore the aggregation, pre-treat cells with 1% DMSO (100µLper 10mL of Essential 8 medium for a 10-cm dish) to improve differentiation of hPS cell lines that are more challenging to differentiate.
Generation of 3D spheroids by using an AggreWell plate
Add 500µLof anti-adherence rinsing solution in a well of a new AggreWell plate and incubate the plate for 0h 5m 0s at 37Room temperature If you notice (under the microscope) bubbles in any of the wells of the plate, centrifuge the AggreWell plate at1300x g in a swinging bucket rotor. Aspirate the rinsing solution and wash with 1mL of PBS.
Pre-warm Essential 8, accutase and DMEM/F12 medium at 37Room temperature. Supplement Essential 8 medium with ROCK inhibitor (Y-27631; 1:1,000) to a final concentration of 10micromolar (µM).
Add 0.5mLof Essential 8 medium supplemented with Y-27632 to each well of an Aggrewell plate. Before starting aggregation, an Aggrewell plate can be centrifuged at 2000x g in a swinging bucket rotor that is fitted with a plate holder to remove any air bubbles in the microwells.
Set the plate aside in an incubator while preparing a single-cell suspension of hPS cells for aggregation.
Aspirate the maintenance medium from the hPS cell culture plates and rinse cells twice with DPBS (without calcium and magnesium).
Add 4mL of accutase per 10-cm plate and incubate at 37°Cfor 0h 7m 0s in a 5% CO2 incubator until cells detach by gentle shaking from the plate.
Gently pipette the cell suspension two to three times with a serological pipette to ensure that any remaining clumps are fully dissociated.
Transfer the cells to a 50mL conical tube.
Add 6mL of DMEM/F12 to rinse any remaining cells off the plate, transfer to a conical tube, and mix the well with the cell suspension in a total volume of 10mL.
Count cells and centrifuge the conical tube containing the cell suspension at 200x g.
Resuspend the cells in Essential 8 medium supplemented with Y-27632 by adjusting the concentration of the single-cell suspension to 2.5–3 million in1mL, and transfer the 1mLof cell suspension into a well of an AggreWell plate to achieve a final volume of 1.5mL per well
.
Pipette gently several times to re-establish an even distribution of cells throughout the well.
Check under a microscope to verify that cells are evenly distributed among the microwells.
Incubate the cells at 37°C, 5% CO2 for 24h 0m 0s.
Dislodging 3D spheroids from the AggreWell plate ● Timing 1 h
Pre-warm DMEM/F12 and Essential 6 medium at 37Room temperature for 0h 30m 0s to1h 0m 0sbefore dislodging.
Place a 40μm cell strainer on top of a 50mL conical tube and pass the suspension of spheroids through the strainer. The aggregates will remain on the filter of the strainer.
Pipette 1mL of DMEM/F-12 medium across the entire surface of the well to dislodge any remaining spheroids. Collect washes and pass over the strainer used in the previous step.
Invert the strainer and place it over a new 50mL conical tube. Collect the spheroids by adding 12mL of Essential 6 medium on top of the inverted strainer. Observe the AggreWell plate under the microscope to ensure that all aggregates have been removed from the wells. Repeat the wash if necessary.
Place spheroids in ultra-low-attachment 10-cm plates in Essential 6 medium supplemented with dorsomorphine (5micromolar (µM)), SB-431542 (10micromolar (µM)).
Neural induction
Add dorsomorphine and SB-431542 to Essential 6 medium for the first 120h 0m 0s. XAV-939 (1.25micromolar (µM)) can also be added for the first 120h 0m 0s of differentiation if forebrain differentiation is not highly efficient.
Perform a medium change daily (but not on day 1) at approximately the same time of the day by gently transferring the neural spheroids to a50mL Falcon tube (if they are too small) and aspirating the medium once they sit at the bottom of the tube. When spheroids are large, perform the medium change by gently tilting the plate (until spheroids settle at the bottom) and removing the remaining medium. This can minimize the transferring time of the spheroids in and out of the plate at this critical differentiation period.
Patterning and differentiation
From day 6, replace the medium daily with NM with supplements listed in option A for hCSs and option B for hStrSs.
TROUBLESHOOTING
(A) Generation of hCSs
From day 6, replace the medium daily with NM supplemented with 20ng/ml FGF2 and 20ng/ml EGF for generating hCSs. Change the medium daily from day 6 to day 15.
From day 16 onward, change the medium every other day until day 22. Continue to use NM supplemented with 20ng/ml FGF2 and 20ng/ml Q29 EGF.
From day 22 to day 46, replace FGF-2 and EGF with 20ng/ml BDNF, 20ng/ml NT-3, 200micromolar (µM) AA, 50micromolar (µM) dibutyryl cAMP sodium salt and 10micromolar (µM) DHA. Perform medium changes with 12mL to 15mL of supplemented NM per 10-cm plate every 2 d.
(B) Generation of hStrSs
From day 6, replace the medium daily with NM supplemented with 50ng/ml activin A and 704 2.5micromolar (µM) IWP-2. Change the medium daily from day 6 to day 11.
From day 12 to day 15, supplement NM with 50ng/ml activin A, 2.5μm IWP-2 and 100nanomolar (nM) SR11237. Perform medium changes with 12mL to 15mL of NM per 10-cm plate every day.
From day 16 onward, change the medium every other day until day 22. Continue to use NM supplemented with 50ng/ml activin A, 2.5μm IWP-2 and 100nanomolar (nM) SR11237. Perform medium changes with 12mL to 15mL of NM per 10-cm plate every 2 d.
From day day 22 to day 41, supplement NM with 20ng/ml BDNF, 20ng/ml NT-3, 200micromolar (µM) AA, 50micromolar (µM) dibutyryl cAMP sodium salt and 10micromolar (µM) DHA. Perform medium changes with 12mL to 15mL of NM per 10-cm plate every 2 d.
From day 42 to day 46, supplement NM with 20ng/ml BDNF, 20ng/ml NT-3, 200micromolar (µM) AA, 50micromolar (µM) dibutyryl cAMP sodium salt, 10micromolar (µM) DHA and 2.5micromolar (µM) DAPT. Perform medium changes with 12 mL to15 mL of NM per 10-cm plate every 2 d.
Long-term culture and phenotyping ● Timing indefinite
After day 46, perform medium changes with 12–15 ml of NM without growth factors every 4–5 d.
(A) Viral labeling of neural spheroids ● Timing 3 d
For labeling neural spheroids, dilute 0.5µL of virus solution (recommended virus particle volumes are listed in Table 1) in 200µL of NM and add mixed NM to a 1.5mL Eppendorf tube. Then, transfer one to three spheroids in the tube containing NM and viruses. The tube can be placed overnight in an incubator at 37°C, 5% CO2.
The next day, add an extra 800µLof fresh NM to the tube and incubate for 24h 0m 0s.
On day 3, wash the spheroids with 1mL of fresh NM (two to three times) and then transfer the spheroids back to their culture plate.
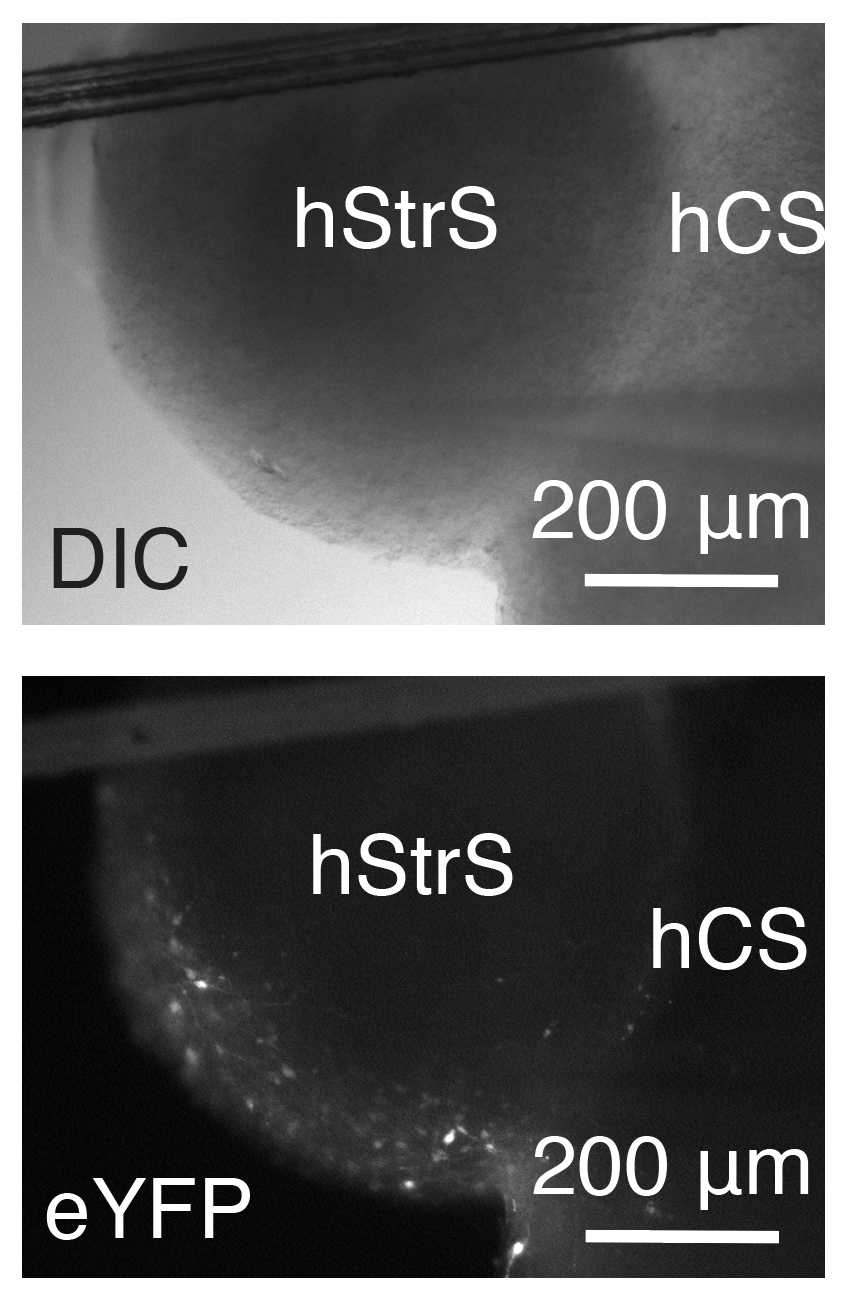
(B) Generating cortico-striatal assembloids ● Timing 3–4 d
To generate cortico-striatal assembloids, transfer one hCS and one hStrS into a 1.5mL Eppendorf tube containing 1mL of NM.
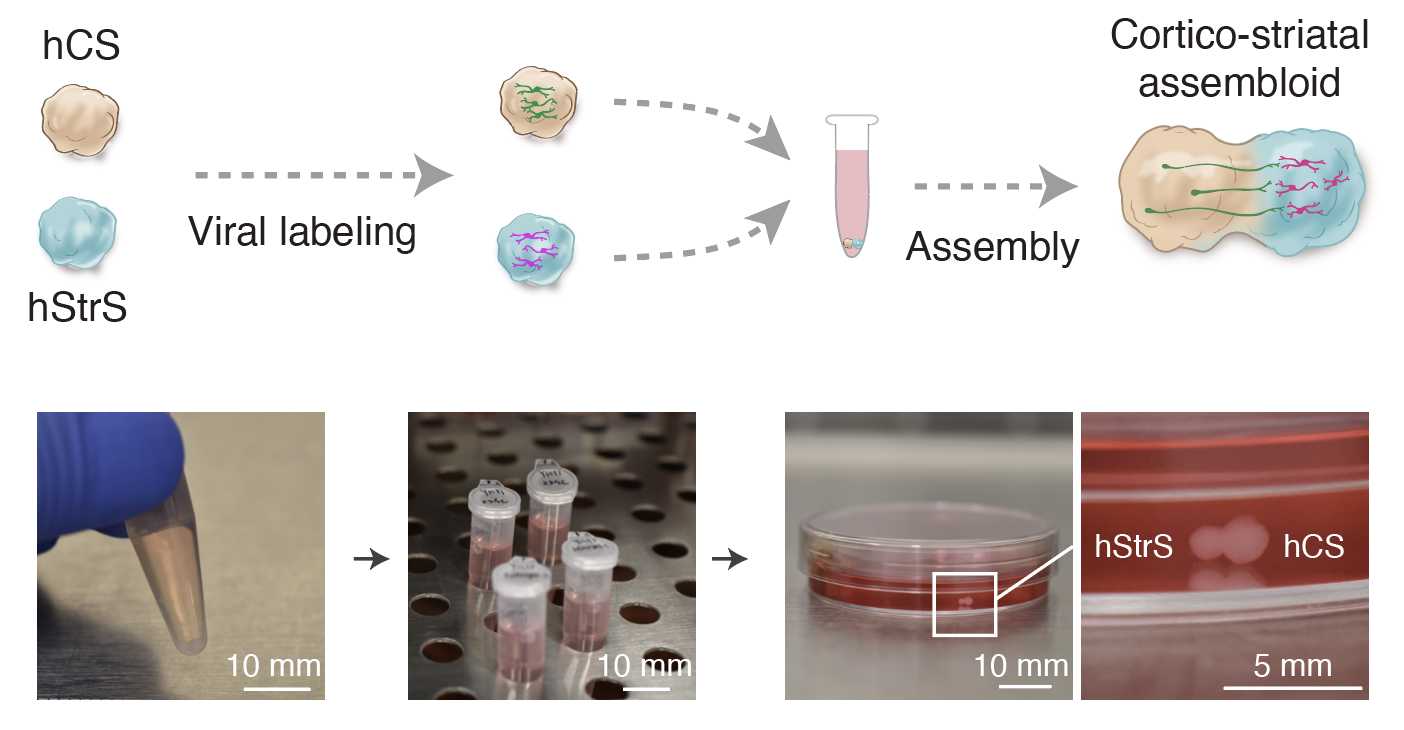
Incubate the spheroids at 37°C for 3–4 d, completely replacing the medium on day 2.
After spheroids are assembled, transfer them into an ultra-low-attachment plate by using a P1000 pipette with the tip cut for a larger bore opening. Assembly of hCS and hStrS between days 46 and 80 is more efficient.
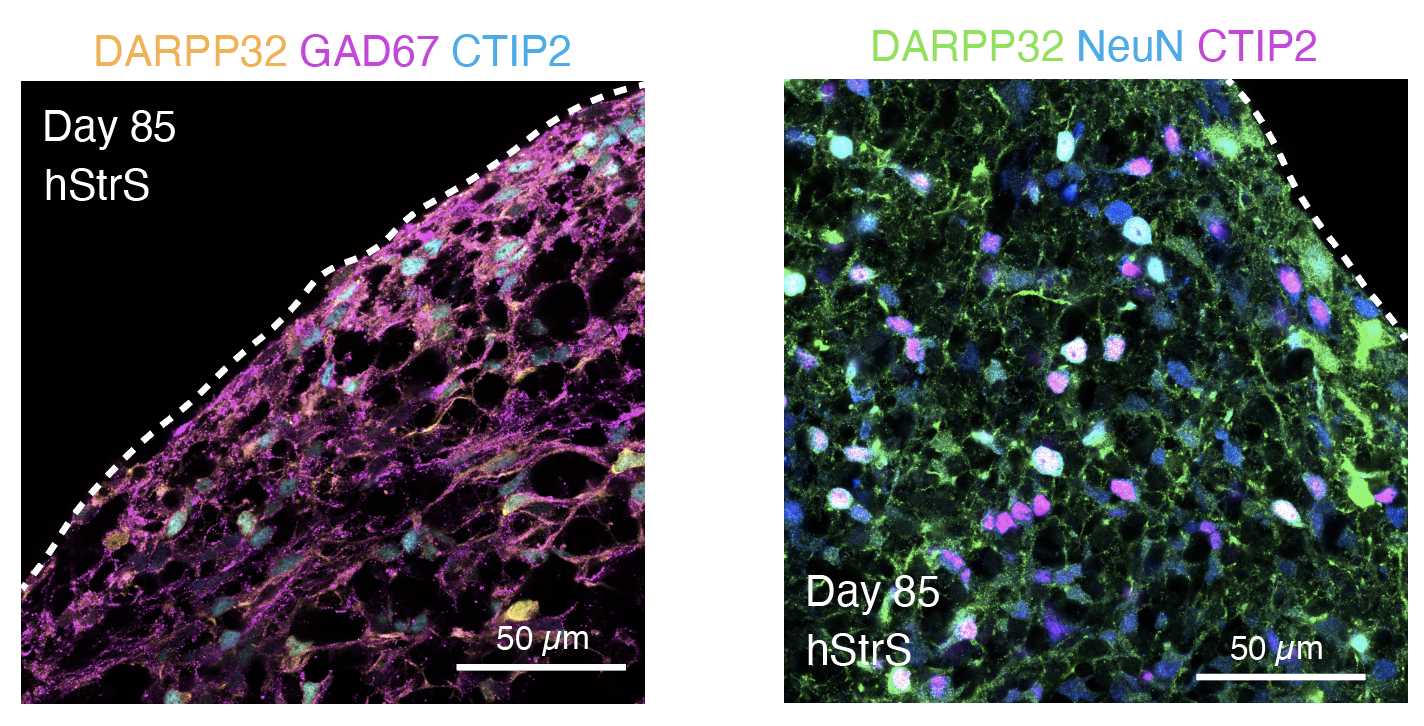
(C) Cryosection and immunostaining ● Timing 6–7 d
When ready to perform immunocytochemistry, transfer spheroids to a 1.5mL microcentrifuge tube by using a cut P1000 tip. Gently remove the medium and add 500µL to 1mL of cold 4% PFA/PBS (or warm 4% PFA/4% sucrose/PBS). Leave spheroids at 4°C 24h 0m 0s.
Collect sections on Superfrost Plus slides. Slides can be stored at -20°C to -80°C until ready to immunostain.
For immunostaining, remove the slide from the freezer and leave at 80Room temperature to thaw for 0h 5m 0s–0h 10m 0s
Wash the slide three times with PBS to remove excess OCT/sucrose on the slides.
Remove PBS and block for 1h 0m 0s at 80Room temperature with 10% normal donkey serum (NDS), 0.3% Triton- X and 0.1% BSA in PBS.
Perform the immunostaining steps in a humidified chamber to avoid drying of sections.
Add the primary antibodies diluted in antibody solution containing 2% NDS and 0.1% Triton-X in PBS and keep at 4°C 1h 0m 0s.
The next day, perform three 0h 5m 0s PBS washes at 4Room temperature (1/3).
The next day, perform three 0h 5m 0s PBS washes at 4Room temperature (2/3).
The next day, perform three 0h 5m 0s PBS washes at 4Room temperature (3/3).
Add secondary antibodies diluted in antibody solution for 1h 0m 0s at 4Room temperature.
After fixation, gently remove PFA and wash with PBS at 4Room temperature.
Perform three 0h 5m 0s washes at 4Room temperature with PBS (1/3).
Perform three 0h 5m 0s washes at 4Room temperature with PBS (2/3).
Perform three 0h 5m 0s washes at 4Room temperature with PBS (3/3).
Add Hoechst in PBS (1:10,000 dilution) for 0h 5m 0s–0h 7m 0s at4Room temperature.
Aspirate the Hoechst solution and wash twice with PBS.
Aspirate PBS and mount the sections by adding one drop of Aquamount solution on top of sections and by placing a coverglass on top.
Use a fluorescent microscope to image.

Add 1mLof 30% (wt/vol) sucrose/PBS for cryopreservation. Leave spheroids or assembloids at 4°C until they sink to the bottom of tube (≥24–48 h).
Prepare square disposable molds that fit with the sample size and number and fill the molds with embedding solution (1:1, OCT/30% sucrose; see Reagent setup).
Transfer spheroids from tubes into the mold by using a P1000 cut pipette tip. Place 4–10 spheroids in the mold. If spheroids move while embedding, use a P20 tip to gently place them in the embedding solution.
Place the spheroids in a cryomold 4On ice for 0h 20m 0s and allow the spheroids to sink to the bottom of the mold.
Snap-freeze the spheroids by placing the mold directly on dry ice. Once completely frozen, store molds at -80°C.
When ready for sectioning, transfer the mold from -80°C to the cryostat chamber at -20°C for ~0h 30m 0s before sectioning.
Remove the block from the mold, paste the specimen on the stage by using OCT and section by using standard techniques. Use a brush to prevent crumbling of the sections. Spheroids and assembloids can be cryo-sectioned at 10 µm –30 µm thickness, depending on the purpose of the experiments.
(D) CUBIC clearing and imaging ● Timing 4–7 d
Transfer spheroids to a 1.5mL microcentrifuge tube by using a cut P1000 tip. Gently remove the medium and add 500µL to 1mL of cold 4% PFA/PBS (or add to 1mL of warm 4% PFA/4% sucrose/PBS and leave at 4Room temperature for 0h 20m 0s). Leave spheroids at 4°C 0h 20m 0s.
Wash with PBS three times at 37°C for 2h 0m 0s (3/3).
Add secondary antibodies diluted in solution containing 0.2% Triton-X100 and 3% NDS in PBS and incubate assembloids at 37°C for 2h 0m 0s.
Wash with PBS three times at 37Room temperature for 2h 0m 0s (1/3).
Wash with PBS three times at 37Room temperature for 2h 0m 0s (2/3).
Wash with PBS three times at 37Room temperature for 2h 0m 0s (3/3).
For refractive index–matching, remove PBS from the 1.5mL tubes, add 1mL of CUBIC-R in the 1.5mL tube and leave for 2h 0m 0s at 37Room temperature.
Once assembloids become transparent, transfer them to a well of a 96-well imaging- compatible plate filled with 150µL of CUBIR-R+ solution and image them with a confocal microscope
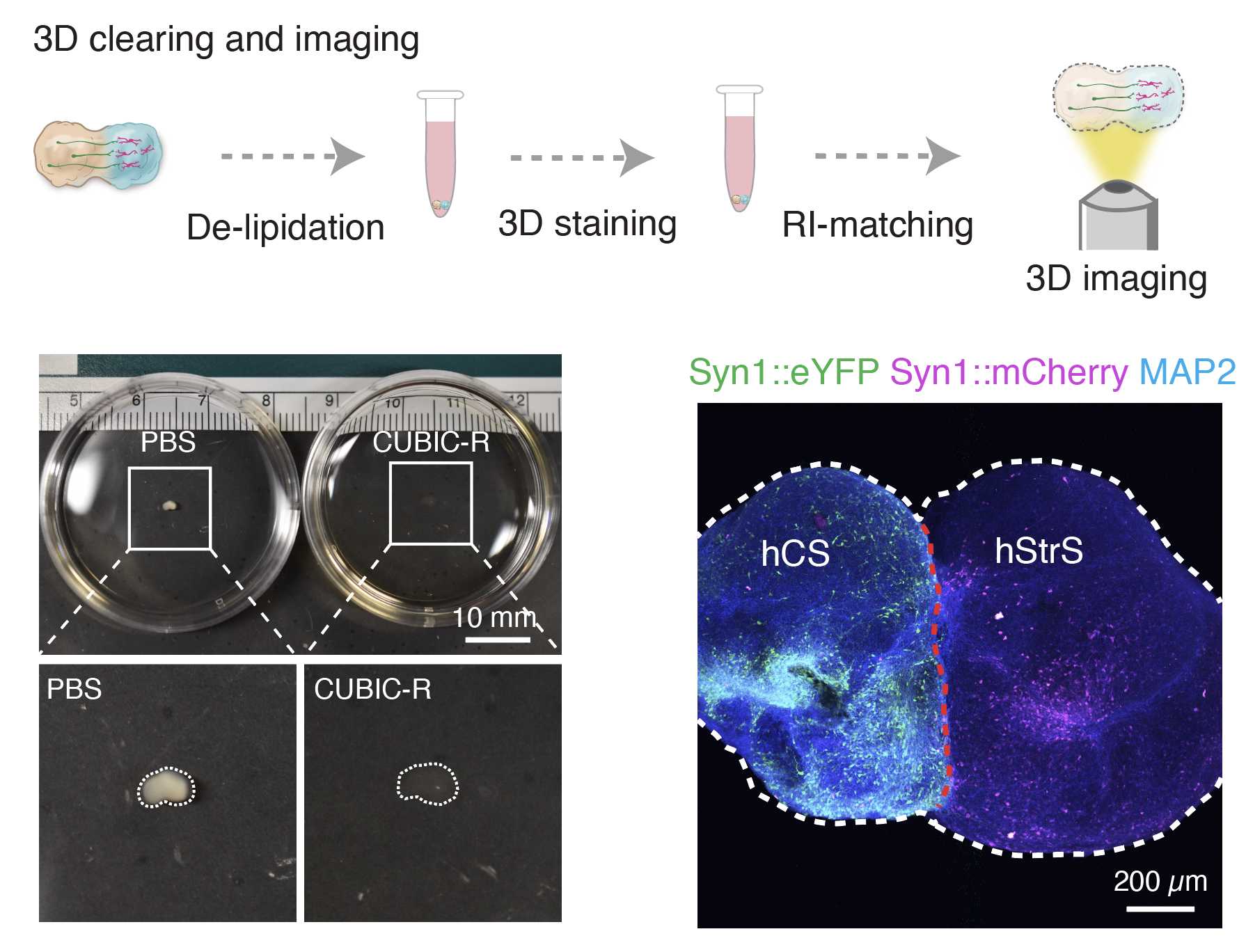
.
After fixation, wash samples with PBS twice at 4Room temperature.
Add 500µL to 1mL of CUBIC-L for de-lipidation and incubate at 37°C for 48h 0m 0s.
Wash with PBS three times at 37Room temperature for 2h 0m 0s (1/3).
Wash with PBS three times at 37Room temperature for 2h 0m 0s (2/3).
Wash with PBS three times at 37Room temperature for 2h 0m 0s (3/3).
Add a combination of primary antibodies diluted in solution containing 0.2% Triton-X100, 3% NDS in PBS and incubate assembloids at 37°C for 2h 0m 0s.
Wash with PBS three times at 37°C for 2h 0m 0s (1/3).
Wash with PBS three times at 37°C for 2h 0m 0s (2/3).
(E) Live imaging of axon growth ● Timing 2–8 h
Using a cut P1000 tip, transfer assembloids to a well of a 96-well imaging-compatible plate in 150µL–200µL of NM.
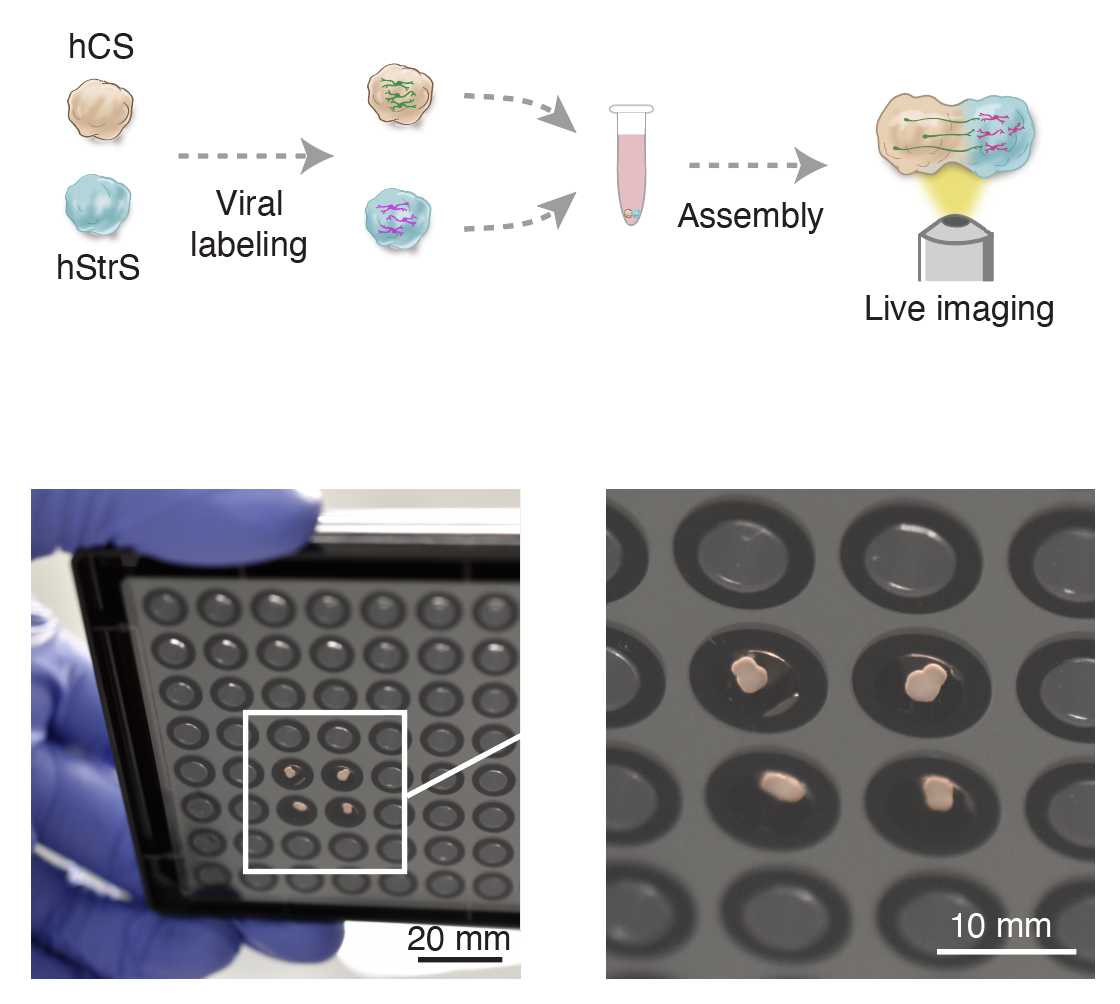
Place the 96-well plate into an inverted microscope with a motorized stage under environmentally controlled conditions (37°C, 5% CO2), and keep the assembloids for 0h 15m 0s–0h 30m 0s before experiments to let assembloids settle at the bottom of the plate.
Set up imaging positions with z-stacks and take images by using an ×10 objective lens at a depth of 0–500 µm.
(F) Optogenetic stimulation and calcium imaging ● Timing 2–6 h
Transfer an assembloid onto a 200 mm glass coverslip in a 35 mm plate in NM and image by using an ×10 objective on a Leica TCS SP8 confocal microscope.
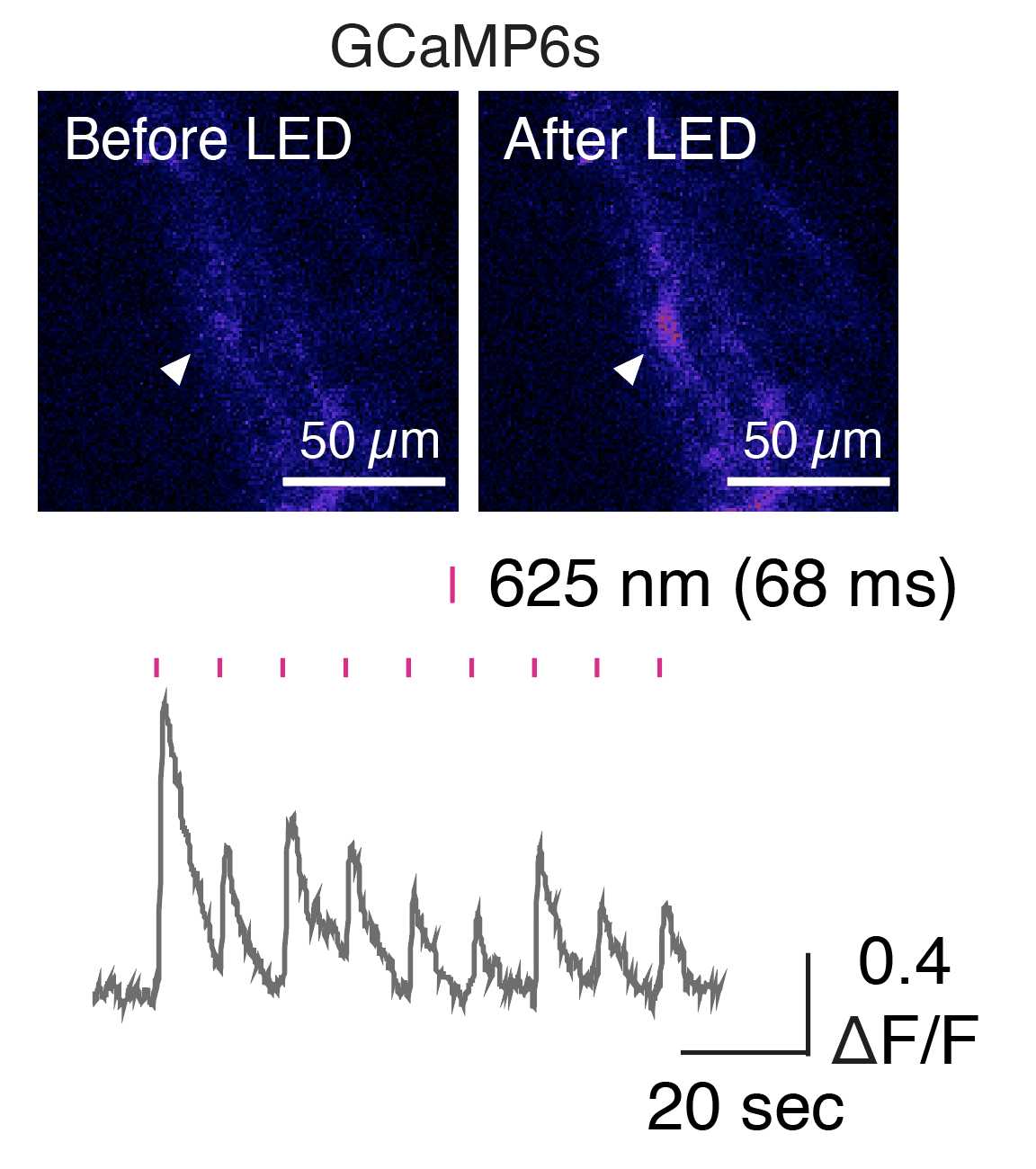
For optogenetic stimulation, activate ChrimsonR-tdTomato+ cells in hCSs with 625 nm light by using an optical fiber-coupled LED. Our stimulation experiments included 1,500 frames, and one 625 nm pulse of LED light (68 ms) was applied every 150 frames as generated by a Cyclops LED connected to the LEICA TCS SP8 microscope.
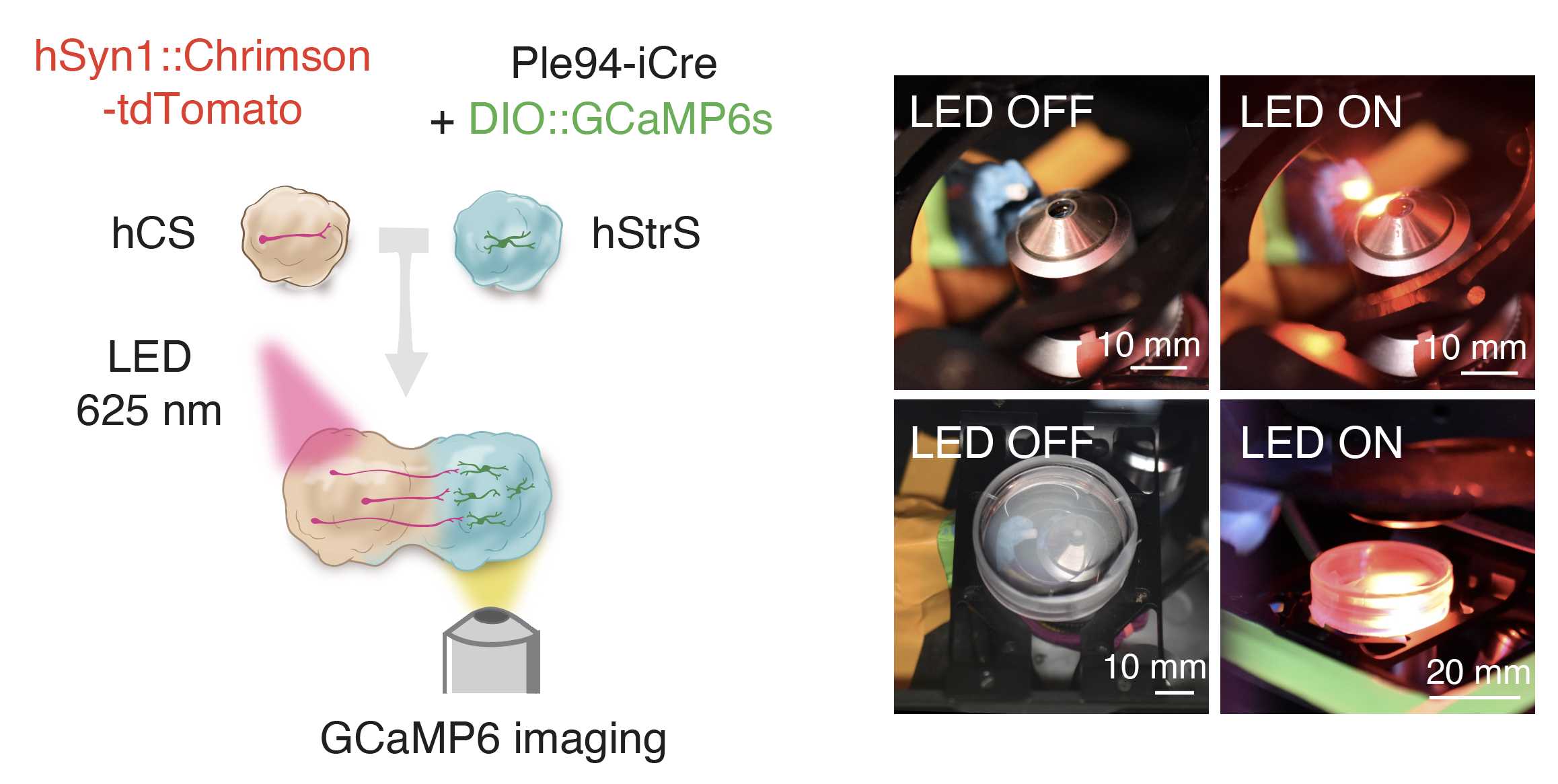
Process raw GCaMP6 signals with Fiji.
After registration of ROIs, transform raw time series to relative changes in fluorescence by using the following formula: ΔF/F = (Ft – F0)/F0.
To verify whether responses are time-locked to LED stimulation, compare ΔF/F responses to ΔF/F values obtained at randomly selected time points.
For quantification of time-locked ΔF/F values, calculate the amplitude of ΔF/F values from each cell as the maximum ΔF/F values within 20–30 frames (1,360–2,040-ms window) after LED stimulation (minus the mean of the baseline 1 s before stimulation).

TROUBLESHOOTING
(G) Whole-cell electrophysiological recording ● Timing 2–6 h
Add 0.2g of low-melt agarose into a 35 mm plate containing 5mLof aCSF and mix well (4% agarose). Dissolve low-melt agarose by using a microwave (avoid spillover due to overheating).
Wait for a few minutes for the agarose to cool down to 37°C and transfer spheroids or assembloids from the medium into agarose with a cut P1000 pipette tip.

Cut a block of agarose out of the culture plate by using a blade and glue the block on the plate of the vibratome. Slice spheroids at a 200 µm to 250 µm thickness with cold aCSF in a vibratome. Transfer slices into aCSF (at 4Room temperature bubbled with 95% O2/5% CO2).
Patch-clamping on spheroid slices is usually performed on fluorescently labeled neurons. Record labeled neurons with glass pipettes with small tips (8–10 MΩ with K-gluconate internal solution, 7–9 MΩ with Cs internal solution)
Once cells are identified through fluorescence, patch under differential interference contrast (DIC) or fluorescence. For DIC, the microscope is switched to a bright field. For patching under fluorescence, there are at least two approaches:
- Increase the bright field exposure under epifluorescence so that both fluorescence (to visualize the cell) and bright field (to visualize the pipette tip) can be seen simultaneously. Once a ‘dimple’ is seen under fluorescence or increased pipette resistance is achieved, release positive pressure and form a tight seal.
Use fluorophore-coated pipettes, patching under epifluorescence. Back-fill glass pipettes by dipping the tips in the BSA-Alexa Fluor (0.02% in PBS) in a 1.5-ml Eppendorf tube for
0h 0m 5sto0h 0m 10sand, after drying for0h 10m 0s, fill pipettes with internal solution and patch.
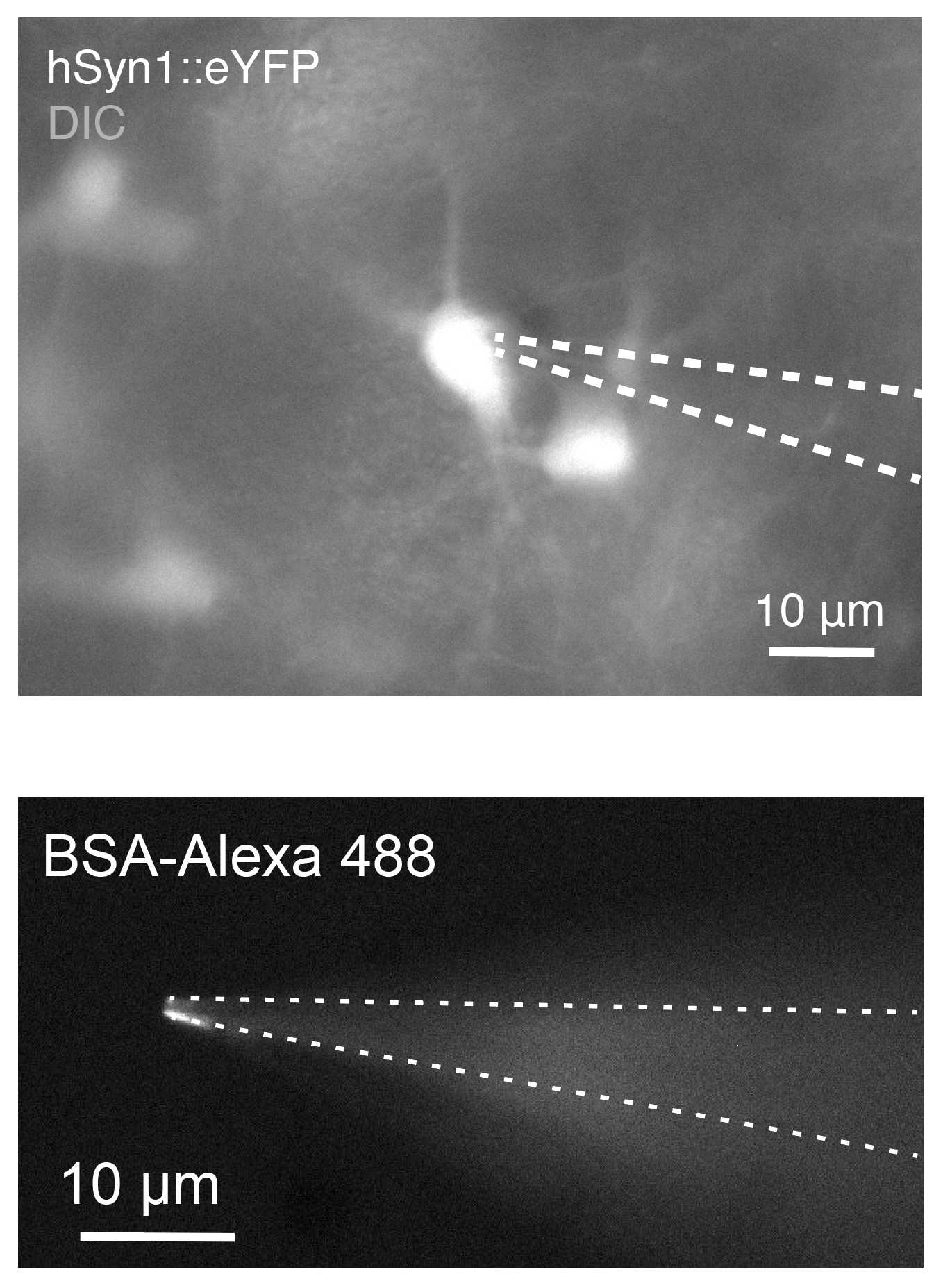
Once the pipette is approaching the target cell, provide minimal positive pressure. Once the tight seal is formed, strong suction is required to break cells.
For recording optically evoked responses in assembloids, apply light at the maximal power through the 40× objective by using a CoolLED (480 nm for ChR2 and 550 nm for Chrimson). CoolLED is controlled by a protocol in pClamp.
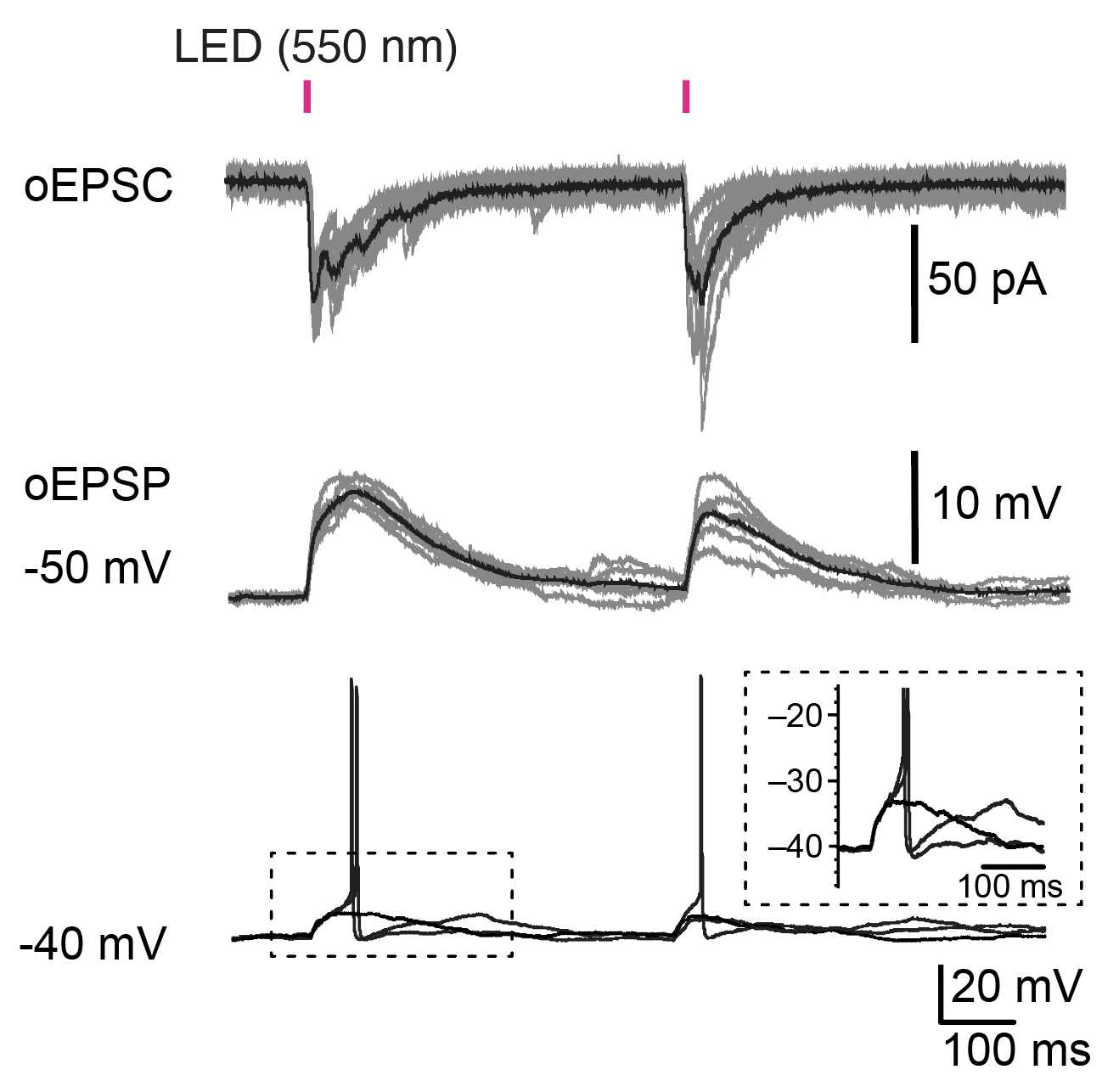
Make recordings at 30°C to 37°Cor37Room temperature within 5h 0m 0s to6h 0m 0safter sectioning.
TROUBLESHOOTING