Analysis of Breast Cancer Cell Invasion Using an Organotypic Culture System
Romana E. Ranftl, Fernando Calvo
Abstract
Metastasis is the main cause of cancer patient mortality. Local tumor invasion is a key step in metastatic dissemination whereby cancer cells dislodge from primary tumors, migrate through the peritumoral stroma and reach the circulation. This is a highly dynamic process occurring in three dimensions that involves interactions between tumor, stromal cells, and the extracellular matrix. Here we describe the organotypic culture system and its utility to study breast cancer cell invasion induced by cancer-associated fibroblasts. This is a three-dimensional model that reproduces the biochemical and physiological properties of real tissue and allows for investigating the molecular and cellular mechanisms involving tumor and its microenvironment, and their contribution to cancer cell invasion. This system provides a robust, accurate, and reproducible method for measuring cancer cell invasion and represents a valuable tool to improve the mechanistic understanding of the initial steps in metastasis.
Before start
This protocol is an adapted version of a previously described method for SCC12 carcinoma cells [10] ( see Fig. 1 for a schematic representation). We have optimized the organotypic invasion assay for murine and human breast cancer cells (410.4/4T1 and MDA-MB-231) in combination with murine or human fibroblasts, respectively ( see Note 8 ). On a general basis, cancer cells are not invasive in this setting and rely on CAF activities to invade. It is recommended to test the behavior of alternative cancer cell types in the assay by plating them on top of a fibroblast-free gel matrix. Heterogeneity is also observed in CAFs in terms of their ability to contract and remodel gel matrices. The amount of fibroblasts and cancer cells to be used, as well as the length of the protocol, need to be optimized if alternative models are used ( see Note 9 ).
Attachments
Steps
3.1 Gel Preparation
Prechill all components for the gel preparation On ice.
Mix 100µL, 80µL , and 120µL.
Add 200µL and 400µL ( see Notes 10 and 11 ). Keep the mixture On ice ( see Note 12 ).
Trypsinize fibroblasts of interest (NFs/CAFs) from a monolayer to a single cell suspension. Count the cells, centrifuge 400x g, and resuspend the pellet at a concentration of 107cells/mL in fibroblast culture medium ( see Notes 13 and 14 ).
Avoiding bubble formation, add 100µL to the gel mix. If a gel without fibroblasts is required, add 100µL instead of cell suspension ( see Note 15 ).
Add 900µL of the mixture in a 24 well-plate well (Fig. 2c) ( see Note 16 ).
Place the plate at 37°C and 5% CO2for 1h 0m 0s.
When the gel is set, add 1mL of appropriate medium on top and incubate 1h 0m 0s at 37°Cand 5% CO2( see Note 17 ).
3.2 Breast Cancer Cell Preparation
Trypsinize a monolayer of breast carcinoma cells (4T1, 410.4 or MBA-MD-231), count the cells and prepare a single cell suspension at 5 × 106cells/mL in cancer cell culture medium.
Aspirate the medium carefully from the 24-well plate containing the gels.
Apply 100µL on the top of each of the gels ( see Notes 18 and 19 ).
Incubate at 37°C and 5% CO2. Leave the cells to adhere to the matrix for 6h 0m 0s–8h 0m 0s.
3.3 Coating of Nylon Filters
Prepare the adequate number of nylon filters (1 per organotypic condition) with a sterile forceps on top of each other in a culture dish.
Prepare 1mL according to the recipe described in Section 3.1 "Gel Preparation."
Coat the nylon filters using 1mL (Fig. 2e).
Separate the coated filters in the culture dish (Fig. 2f).
Incubate the coated nylon filters at 37°C for 1h 0m 0s.
Fix the coated nylon, using filter fixing solution for at least 2h 0m 0s at 37Room temperature or at 4°C.
3.4 Lifting the Gel and Covering the Cancer Cells
Wash the gel-coated nylon filters with sterile PBS for 0h 10m 0s (×3) to remove all traces of fixing solution.
Wash the gel-coated nylon filters with sterile PBS for 0h 10m 0s (1/3)
Wash the gel-coated nylon filters with sterile PBS for 0h 10m 0s (2/3)
Wash the gel-coated nylon filters with sterile PBS for 0h 10m 0s (3/3)
Add cancer cell culture medium to the filters and incubate for 0h 30m 0s at 37°C.
Place the sterile metal grid bridges in a 6-well plate with sterile forceps (Fig. 2g).
Add a coated, washed and medium-adapted nylon filter on top of each metal bridge with sterile forceps (Fig. 2h).
Prepare an appropriate amount of cell-free gel according to the recipe in Section "Gel Preparation" and keep the mixture 37On ice (100µL per organotypic gel).
Remove the medium on the top of the gels carefully in order not to disturb the organotypic cultures.
Lift the gels with a sterile spatula from the 24-well and place it on a coated filter on top of a metal bridge. The cancer cell layer must be facing up (Fig. 2i, j).
Add medium under the bridge until it is in contact with the nylon filter. Avoid air bubbles between the interphase medium/nylon filter (Fig. 2k).
Add 100µL on top of the lifted gels and spread over the surface.
Incubate the culture for 5 days (37°C, 5% CO2) and change the medium daily ( see Note 21 ).
3.5 Gel Fixing and Processing
After culturing for 5 days ( see Note 9 ), remove the medium from the plate and wash with PBS or transfer the organotypic cultures to a new 6-well plate by lifting the filter and gel. Turn the gel upside-down for fixing (cancer cell layer on the bottom, Fig. 1) ( see Note 22 ).
Fix the gels in 2mLat 4°C 0h 30m 0s.
Next day, wash the gels with 2mL for 0h 10m 0s (×3).
Wash the gels with 2mL for 0h 10m 0s (1/3)
Wash the gels with 2mL for 0h 10m 0s (2/3)
Wash the gels with 2mL for 0h 10m 0s (3/3)
Using the forceps and the scalpel cut the gel in two halves and store one of them in 70% ethanol at 4°C ( see Note 23 ).
Embed the other half of the gel in paraffin blocks and perform standard H&E staining on sections ( see Notes 24 and 25 ).
3.6 Data Analysis
Take 5–7 pictures per gel using a bright field microscope (20×/10× magnification) ( see Note 26 and Fig. 3a-e).
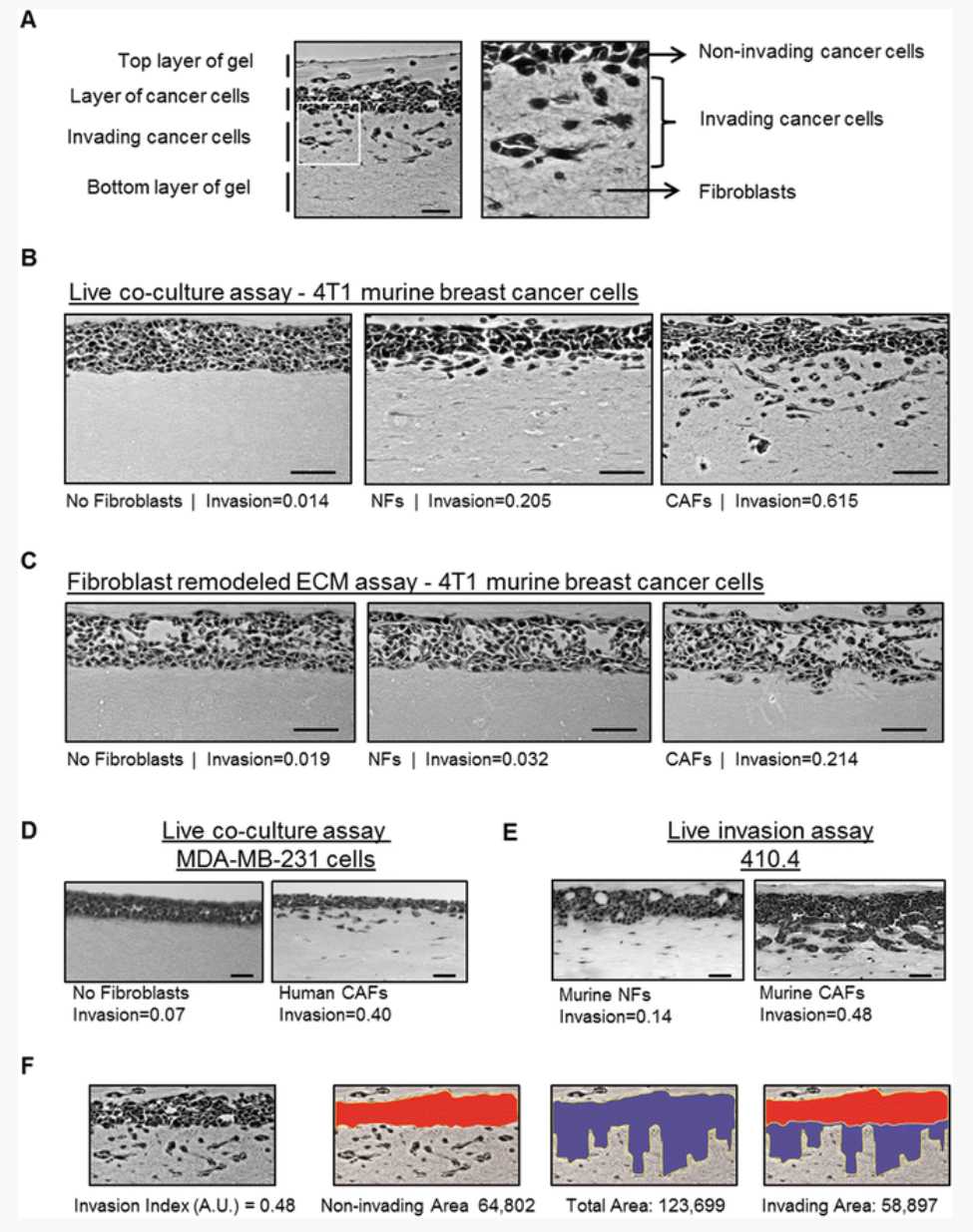
Measure the area of noninvasive cancer cells and the total area of cancer cells (noninvasive and invasive) using Image J or a similar software (Fig. 3f).
The Invasion Index is calculated by dividing the invading area (total area − invading area) by the total area ( see Note 27 ).
3.7 Killing Assay
Follow the protocol as described in Section 3.1 "Gel Preparation" ( steps 2 –9). Once the gel is set, add 1mL on top.
Incubate at 37°C, 5% CO2for 5 days. Change the medium daily ( see Notes 17 and 28 ).
Fibroblast removal (“killing” step): Remove the medium from the gels and add 1mLcontaining an appropriate selection compound (e.g., 10 μg/mL puromycin) ( see Note 29 ).
Incubate at 37°C, 5% CO2for 48h 0m 0s.
Wash the gels for 1h 0m 0s in fibroblast culture medium (×3).
Wash the gels for 1h 0m 0s in fibroblast culture medium (1/3)
Wash the gels for 1h 0m 0s in fibroblast culture medium (2/3)
Wash the gels for 1h 0m 0s in fibroblast culture medium (3/3)
Incubate at 37°C, 5% CO21h 0m 0s in fibroblast culture medium to remove all traces of selection compound ( see Note 30 ).
Continue with the procedure as described in Sections 3.2 "Breast Cancer Cell Preparation" – 3.6 "Data Analysis".

